Overall, Pycaret is an incredibly useful and powerful library for speeding up and automating the machine learning pipeline and process. Lets highlight some key pros and cons.
Pros
Less code: The library really lives up to its motto of being a ‘low code’ library, often one of code will replace what would normally have been an entire manually coded process of many lines of code. Accross a whole project, as we have seen in this example project, hundreds of lines of code can be replace by just a few lines. Note how most of this article length is more due to describing what the code does, than the code itself!
Easy to use: Pycaret library functions are well named, intiutive and easy to use, and easy to customise and configure.
A more consistant approach: Another benefit of being a low code library where most processes have been automated is that this ensures a more consistant approach when using Pycaret accross different projects. This is important not only for scientific reproducability, but for reducing the possibility of errors that are more likely when more custom and manual code is required to be written for a process.
Good practice: Each step of the machine learning pipeline that Pycaret simplifies and automates for you, does so in such a way to bake in best practice in Data Science. For example, when testing models cross-fold validation is done by default on all models. When evaluating models, multiple and relevant metrics are used to evaluate performance.
Performs all key tasks and more: Pycaret automates every key task in machine learning process, from wrangling to preparing your data, for selecting a model, for optimising and evaluating a final model, then testing and saving a model ready for deployment and use. In addition, Pycaret offers easy access to extra functions while not always required, can be useful for particular projects - for example the ability to calculate Shapley values as we have seen for model interpretability.
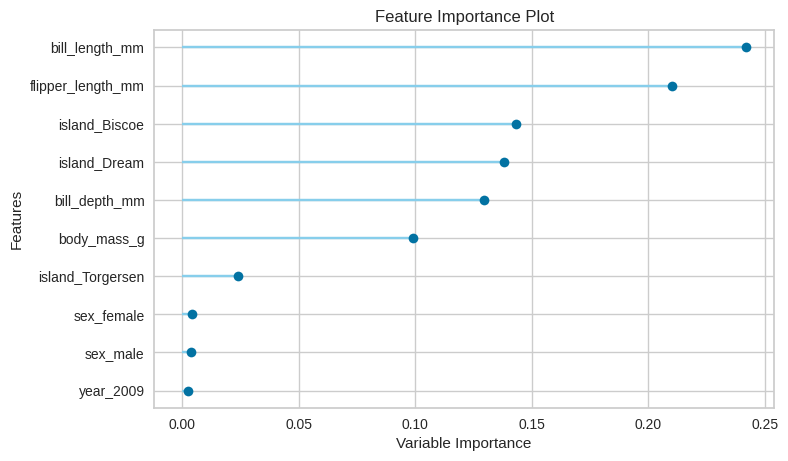
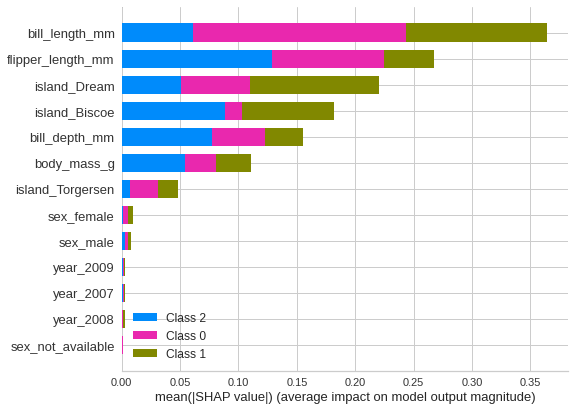
Educational: Using this library helps all kinds of users, from amateurs to professional Data Scientists, keep up to date with the latest methods and techniques. For example, Pycaret maintains a list of the most widely used models which are included automatically when selecting a potential model. For model understanding and interpretation, a wide range of plots and analyses are available. I was not fully aware for example about Shapley values, and how they can help interpret models from a very different perspective. These are some of the many advantages of having an open source library like Pycaret that’s intended to automate the Data Science process, everyone’s collaberative effort to use and update the library helps keep highlighting and providing some of the latest and best techniques to all who use it.
Excellent data wrangling and transformation: As we saw with the setup() function there are many useful features available to perform many common tasks that would normally require many lines of code. For example, the inclusion of the SMOTE and resampling techniques often used to correct for imbalances in the target variable in a dataset. Sensible automatic imputation methods by default to deal with missing values, and normalisation methods to scale and prepare numeric data - are key common tasks that need to be performed, expertly automated by the Pycaret library.
Quick consideration of a wide range of models: Pycaret’s compare_models(), create_model() and tune_model() functions allow you to quickly compare a wide range of the best models available (currently 18), then select and optimise the best model - in just 3 lines of code.
Creating a pipeline not just a model: The machine learning process is not just about producing a good model, you also need a process to transform the data into a format required for that model. This is often consider a separate bit of extra work, often referred to as an ETL process. (Extract, Transform & Load). Pycaret blends these two essential things together for you, another benefit of the automation it provides, so when you save your model, you also save this data transformation process, all together. And when you load it ready for use, you load the data transformation and the model together - ready for immediate use - a huge saving of time and effort.
These are just some of the key pros of the Pycaret library, in my opinion there are many many more. To illustrate what a huge advance and benefit the Pycaret library is in the pros highlighted, compare this to a previous machine learning project of mine to classify breast cancer data, where I used the common and more manual process of many more lines of code for each part of the machine learning pipeline.
Cons
Not good for beginners: Despite being pitched for begginners, this library may not be ideal for beginners in my opinion. While the functions are easy for a beginner to use, and indeed as highlighted you can run the entire machine learning process very easily, I would say this can be a bit deceptive and misleading. Simply running the process with little understanding what is going on underneath, is not a substitute for understanding the basics. For example when, why & how should we transform data? (e.g. normalisation of numeric values) which is the most appropriate metric to interpret results? (e.g. balanced vs imbalanced target variable).
No ability to customose plots: This is perhaps a minor issue, but it would be nice to be able to customise plots at least a little for example to adjust the size of plots.
Can’t easily see what is going on under the hood: In a way, this is I feel both a Pro and a Con. If you know what is going on with these automated functions underneath, then to some extent it can be nice to not be overloaded with lots of detail about it. On the other hand, for both experienced Data Scientist’s and begginners it can be helpful to actually understand more of what each automated function is doing. Many functions do give some insight as to what they are doing, but many things are hidden - and can only be discovered by reading the documentation, which I would suggest is a good idea for anyone using this library, experienced or not. But again I feel this is a relatively minor con, as its a difficult balance to achieve in the trade off between simplifying and automating the process vs making every part of the process transparent.