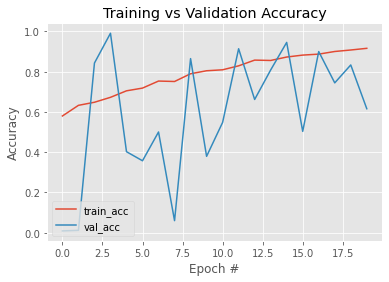
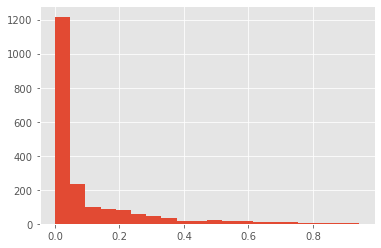
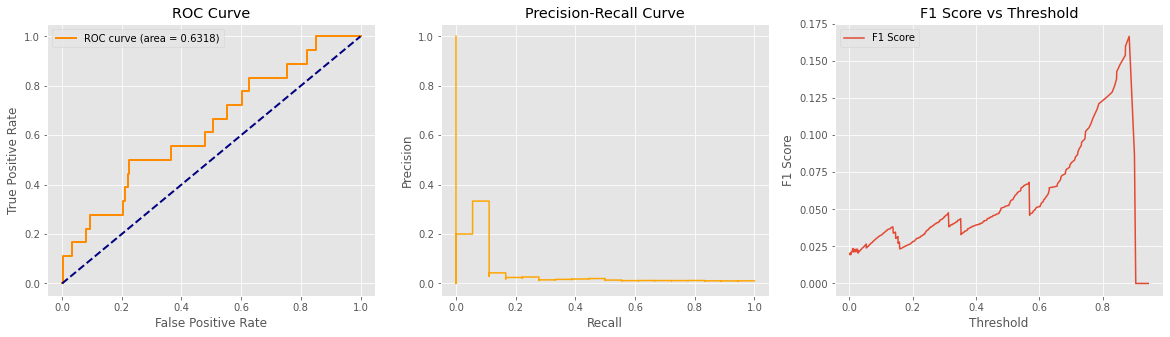
Downloading data from https://github.com/keras-team/keras-applications/releases/download/resnet/resnet50_weights_tf_dim_ordering_tf_kernels.h5
102973440/102967424 [==============================] - 1s 0us/step
Model: "model_1"
__________________________________________________________________________________________________
Layer (type) Output Shape Param # Connected to
==================================================================================================
input_1 (InputLayer) (None, 224, 224, 3) 0
__________________________________________________________________________________________________
conv1_pad (ZeroPadding2D) (None, 230, 230, 3) 0 input_1[0][0]
__________________________________________________________________________________________________
conv1_conv (Conv2D) (None, 112, 112, 64) 9472 conv1_pad[0][0]
__________________________________________________________________________________________________
conv1_bn (BatchNormalization) (None, 112, 112, 64) 256 conv1_conv[0][0]
__________________________________________________________________________________________________
conv1_relu (Activation) (None, 112, 112, 64) 0 conv1_bn[0][0]
__________________________________________________________________________________________________
pool1_pad (ZeroPadding2D) (None, 114, 114, 64) 0 conv1_relu[0][0]
__________________________________________________________________________________________________
pool1_pool (MaxPooling2D) (None, 56, 56, 64) 0 pool1_pad[0][0]
__________________________________________________________________________________________________
conv2_block1_1_conv (Conv2D) (None, 56, 56, 64) 4160 pool1_pool[0][0]
__________________________________________________________________________________________________
conv2_block1_1_bn (BatchNormali (None, 56, 56, 64) 256 conv2_block1_1_conv[0][0]
__________________________________________________________________________________________________
conv2_block1_1_relu (Activation (None, 56, 56, 64) 0 conv2_block1_1_bn[0][0]
__________________________________________________________________________________________________
conv2_block1_2_conv (Conv2D) (None, 56, 56, 64) 36928 conv2_block1_1_relu[0][0]
__________________________________________________________________________________________________
conv2_block1_2_bn (BatchNormali (None, 56, 56, 64) 256 conv2_block1_2_conv[0][0]
__________________________________________________________________________________________________
conv2_block1_2_relu (Activation (None, 56, 56, 64) 0 conv2_block1_2_bn[0][0]
__________________________________________________________________________________________________
conv2_block1_0_conv (Conv2D) (None, 56, 56, 256) 16640 pool1_pool[0][0]
__________________________________________________________________________________________________
conv2_block1_3_conv (Conv2D) (None, 56, 56, 256) 16640 conv2_block1_2_relu[0][0]
__________________________________________________________________________________________________
conv2_block1_0_bn (BatchNormali (None, 56, 56, 256) 1024 conv2_block1_0_conv[0][0]
__________________________________________________________________________________________________
conv2_block1_3_bn (BatchNormali (None, 56, 56, 256) 1024 conv2_block1_3_conv[0][0]
__________________________________________________________________________________________________
conv2_block1_add (Add) (None, 56, 56, 256) 0 conv2_block1_0_bn[0][0]
conv2_block1_3_bn[0][0]
__________________________________________________________________________________________________
conv2_block1_out (Activation) (None, 56, 56, 256) 0 conv2_block1_add[0][0]
__________________________________________________________________________________________________
conv2_block2_1_conv (Conv2D) (None, 56, 56, 64) 16448 conv2_block1_out[0][0]
__________________________________________________________________________________________________
conv2_block2_1_bn (BatchNormali (None, 56, 56, 64) 256 conv2_block2_1_conv[0][0]
__________________________________________________________________________________________________
conv2_block2_1_relu (Activation (None, 56, 56, 64) 0 conv2_block2_1_bn[0][0]
__________________________________________________________________________________________________
conv2_block2_2_conv (Conv2D) (None, 56, 56, 64) 36928 conv2_block2_1_relu[0][0]
__________________________________________________________________________________________________
conv2_block2_2_bn (BatchNormali (None, 56, 56, 64) 256 conv2_block2_2_conv[0][0]
__________________________________________________________________________________________________
conv2_block2_2_relu (Activation (None, 56, 56, 64) 0 conv2_block2_2_bn[0][0]
__________________________________________________________________________________________________
conv2_block2_3_conv (Conv2D) (None, 56, 56, 256) 16640 conv2_block2_2_relu[0][0]
__________________________________________________________________________________________________
conv2_block2_3_bn (BatchNormali (None, 56, 56, 256) 1024 conv2_block2_3_conv[0][0]
__________________________________________________________________________________________________
conv2_block2_add (Add) (None, 56, 56, 256) 0 conv2_block1_out[0][0]
conv2_block2_3_bn[0][0]
__________________________________________________________________________________________________
conv2_block2_out (Activation) (None, 56, 56, 256) 0 conv2_block2_add[0][0]
__________________________________________________________________________________________________
conv2_block3_1_conv (Conv2D) (None, 56, 56, 64) 16448 conv2_block2_out[0][0]
__________________________________________________________________________________________________
conv2_block3_1_bn (BatchNormali (None, 56, 56, 64) 256 conv2_block3_1_conv[0][0]
__________________________________________________________________________________________________
conv2_block3_1_relu (Activation (None, 56, 56, 64) 0 conv2_block3_1_bn[0][0]
__________________________________________________________________________________________________
conv2_block3_2_conv (Conv2D) (None, 56, 56, 64) 36928 conv2_block3_1_relu[0][0]
__________________________________________________________________________________________________
conv2_block3_2_bn (BatchNormali (None, 56, 56, 64) 256 conv2_block3_2_conv[0][0]
__________________________________________________________________________________________________
conv2_block3_2_relu (Activation (None, 56, 56, 64) 0 conv2_block3_2_bn[0][0]
__________________________________________________________________________________________________
conv2_block3_3_conv (Conv2D) (None, 56, 56, 256) 16640 conv2_block3_2_relu[0][0]
__________________________________________________________________________________________________
conv2_block3_3_bn (BatchNormali (None, 56, 56, 256) 1024 conv2_block3_3_conv[0][0]
__________________________________________________________________________________________________
conv2_block3_add (Add) (None, 56, 56, 256) 0 conv2_block2_out[0][0]
conv2_block3_3_bn[0][0]
__________________________________________________________________________________________________
conv2_block3_out (Activation) (None, 56, 56, 256) 0 conv2_block3_add[0][0]
__________________________________________________________________________________________________
conv3_block1_1_conv (Conv2D) (None, 28, 28, 128) 32896 conv2_block3_out[0][0]
__________________________________________________________________________________________________
conv3_block1_1_bn (BatchNormali (None, 28, 28, 128) 512 conv3_block1_1_conv[0][0]
__________________________________________________________________________________________________
conv3_block1_1_relu (Activation (None, 28, 28, 128) 0 conv3_block1_1_bn[0][0]
__________________________________________________________________________________________________
conv3_block1_2_conv (Conv2D) (None, 28, 28, 128) 147584 conv3_block1_1_relu[0][0]
__________________________________________________________________________________________________
conv3_block1_2_bn (BatchNormali (None, 28, 28, 128) 512 conv3_block1_2_conv[0][0]
__________________________________________________________________________________________________
conv3_block1_2_relu (Activation (None, 28, 28, 128) 0 conv3_block1_2_bn[0][0]
__________________________________________________________________________________________________
conv3_block1_0_conv (Conv2D) (None, 28, 28, 512) 131584 conv2_block3_out[0][0]
__________________________________________________________________________________________________
conv3_block1_3_conv (Conv2D) (None, 28, 28, 512) 66048 conv3_block1_2_relu[0][0]
__________________________________________________________________________________________________
conv3_block1_0_bn (BatchNormali (None, 28, 28, 512) 2048 conv3_block1_0_conv[0][0]
__________________________________________________________________________________________________
conv3_block1_3_bn (BatchNormali (None, 28, 28, 512) 2048 conv3_block1_3_conv[0][0]
__________________________________________________________________________________________________
conv3_block1_add (Add) (None, 28, 28, 512) 0 conv3_block1_0_bn[0][0]
conv3_block1_3_bn[0][0]
__________________________________________________________________________________________________
conv3_block1_out (Activation) (None, 28, 28, 512) 0 conv3_block1_add[0][0]
__________________________________________________________________________________________________
conv3_block2_1_conv (Conv2D) (None, 28, 28, 128) 65664 conv3_block1_out[0][0]
__________________________________________________________________________________________________
conv3_block2_1_bn (BatchNormali (None, 28, 28, 128) 512 conv3_block2_1_conv[0][0]
__________________________________________________________________________________________________
conv3_block2_1_relu (Activation (None, 28, 28, 128) 0 conv3_block2_1_bn[0][0]
__________________________________________________________________________________________________
conv3_block2_2_conv (Conv2D) (None, 28, 28, 128) 147584 conv3_block2_1_relu[0][0]
__________________________________________________________________________________________________
conv3_block2_2_bn (BatchNormali (None, 28, 28, 128) 512 conv3_block2_2_conv[0][0]
__________________________________________________________________________________________________
conv3_block2_2_relu (Activation (None, 28, 28, 128) 0 conv3_block2_2_bn[0][0]
__________________________________________________________________________________________________
conv3_block2_3_conv (Conv2D) (None, 28, 28, 512) 66048 conv3_block2_2_relu[0][0]
__________________________________________________________________________________________________
conv3_block2_3_bn (BatchNormali (None, 28, 28, 512) 2048 conv3_block2_3_conv[0][0]
__________________________________________________________________________________________________
conv3_block2_add (Add) (None, 28, 28, 512) 0 conv3_block1_out[0][0]
conv3_block2_3_bn[0][0]
__________________________________________________________________________________________________
conv3_block2_out (Activation) (None, 28, 28, 512) 0 conv3_block2_add[0][0]
__________________________________________________________________________________________________
conv3_block3_1_conv (Conv2D) (None, 28, 28, 128) 65664 conv3_block2_out[0][0]
__________________________________________________________________________________________________
conv3_block3_1_bn (BatchNormali (None, 28, 28, 128) 512 conv3_block3_1_conv[0][0]
__________________________________________________________________________________________________
conv3_block3_1_relu (Activation (None, 28, 28, 128) 0 conv3_block3_1_bn[0][0]
__________________________________________________________________________________________________
conv3_block3_2_conv (Conv2D) (None, 28, 28, 128) 147584 conv3_block3_1_relu[0][0]
__________________________________________________________________________________________________
conv3_block3_2_bn (BatchNormali (None, 28, 28, 128) 512 conv3_block3_2_conv[0][0]
__________________________________________________________________________________________________
conv3_block3_2_relu (Activation (None, 28, 28, 128) 0 conv3_block3_2_bn[0][0]
__________________________________________________________________________________________________
conv3_block3_3_conv (Conv2D) (None, 28, 28, 512) 66048 conv3_block3_2_relu[0][0]
__________________________________________________________________________________________________
conv3_block3_3_bn (BatchNormali (None, 28, 28, 512) 2048 conv3_block3_3_conv[0][0]
__________________________________________________________________________________________________
conv3_block3_add (Add) (None, 28, 28, 512) 0 conv3_block2_out[0][0]
conv3_block3_3_bn[0][0]
__________________________________________________________________________________________________
conv3_block3_out (Activation) (None, 28, 28, 512) 0 conv3_block3_add[0][0]
__________________________________________________________________________________________________
conv3_block4_1_conv (Conv2D) (None, 28, 28, 128) 65664 conv3_block3_out[0][0]
__________________________________________________________________________________________________
conv3_block4_1_bn (BatchNormali (None, 28, 28, 128) 512 conv3_block4_1_conv[0][0]
__________________________________________________________________________________________________
conv3_block4_1_relu (Activation (None, 28, 28, 128) 0 conv3_block4_1_bn[0][0]
__________________________________________________________________________________________________
conv3_block4_2_conv (Conv2D) (None, 28, 28, 128) 147584 conv3_block4_1_relu[0][0]
__________________________________________________________________________________________________
conv3_block4_2_bn (BatchNormali (None, 28, 28, 128) 512 conv3_block4_2_conv[0][0]
__________________________________________________________________________________________________
conv3_block4_2_relu (Activation (None, 28, 28, 128) 0 conv3_block4_2_bn[0][0]
__________________________________________________________________________________________________
conv3_block4_3_conv (Conv2D) (None, 28, 28, 512) 66048 conv3_block4_2_relu[0][0]
__________________________________________________________________________________________________
conv3_block4_3_bn (BatchNormali (None, 28, 28, 512) 2048 conv3_block4_3_conv[0][0]
__________________________________________________________________________________________________
conv3_block4_add (Add) (None, 28, 28, 512) 0 conv3_block3_out[0][0]
conv3_block4_3_bn[0][0]
__________________________________________________________________________________________________
conv3_block4_out (Activation) (None, 28, 28, 512) 0 conv3_block4_add[0][0]
__________________________________________________________________________________________________
conv4_block1_1_conv (Conv2D) (None, 14, 14, 256) 131328 conv3_block4_out[0][0]
__________________________________________________________________________________________________
conv4_block1_1_bn (BatchNormali (None, 14, 14, 256) 1024 conv4_block1_1_conv[0][0]
__________________________________________________________________________________________________
conv4_block1_1_relu (Activation (None, 14, 14, 256) 0 conv4_block1_1_bn[0][0]
__________________________________________________________________________________________________
conv4_block1_2_conv (Conv2D) (None, 14, 14, 256) 590080 conv4_block1_1_relu[0][0]
__________________________________________________________________________________________________
conv4_block1_2_bn (BatchNormali (None, 14, 14, 256) 1024 conv4_block1_2_conv[0][0]
__________________________________________________________________________________________________
conv4_block1_2_relu (Activation (None, 14, 14, 256) 0 conv4_block1_2_bn[0][0]
__________________________________________________________________________________________________
conv4_block1_0_conv (Conv2D) (None, 14, 14, 1024) 525312 conv3_block4_out[0][0]
__________________________________________________________________________________________________
conv4_block1_3_conv (Conv2D) (None, 14, 14, 1024) 263168 conv4_block1_2_relu[0][0]
__________________________________________________________________________________________________
conv4_block1_0_bn (BatchNormali (None, 14, 14, 1024) 4096 conv4_block1_0_conv[0][0]
__________________________________________________________________________________________________
conv4_block1_3_bn (BatchNormali (None, 14, 14, 1024) 4096 conv4_block1_3_conv[0][0]
__________________________________________________________________________________________________
conv4_block1_add (Add) (None, 14, 14, 1024) 0 conv4_block1_0_bn[0][0]
conv4_block1_3_bn[0][0]
__________________________________________________________________________________________________
conv4_block1_out (Activation) (None, 14, 14, 1024) 0 conv4_block1_add[0][0]
__________________________________________________________________________________________________
conv4_block2_1_conv (Conv2D) (None, 14, 14, 256) 262400 conv4_block1_out[0][0]
__________________________________________________________________________________________________
conv4_block2_1_bn (BatchNormali (None, 14, 14, 256) 1024 conv4_block2_1_conv[0][0]
__________________________________________________________________________________________________
conv4_block2_1_relu (Activation (None, 14, 14, 256) 0 conv4_block2_1_bn[0][0]
__________________________________________________________________________________________________
conv4_block2_2_conv (Conv2D) (None, 14, 14, 256) 590080 conv4_block2_1_relu[0][0]
__________________________________________________________________________________________________
conv4_block2_2_bn (BatchNormali (None, 14, 14, 256) 1024 conv4_block2_2_conv[0][0]
__________________________________________________________________________________________________
conv4_block2_2_relu (Activation (None, 14, 14, 256) 0 conv4_block2_2_bn[0][0]
__________________________________________________________________________________________________
conv4_block2_3_conv (Conv2D) (None, 14, 14, 1024) 263168 conv4_block2_2_relu[0][0]
__________________________________________________________________________________________________
conv4_block2_3_bn (BatchNormali (None, 14, 14, 1024) 4096 conv4_block2_3_conv[0][0]
__________________________________________________________________________________________________
conv4_block2_add (Add) (None, 14, 14, 1024) 0 conv4_block1_out[0][0]
conv4_block2_3_bn[0][0]
__________________________________________________________________________________________________
conv4_block2_out (Activation) (None, 14, 14, 1024) 0 conv4_block2_add[0][0]
__________________________________________________________________________________________________
conv4_block3_1_conv (Conv2D) (None, 14, 14, 256) 262400 conv4_block2_out[0][0]
__________________________________________________________________________________________________
conv4_block3_1_bn (BatchNormali (None, 14, 14, 256) 1024 conv4_block3_1_conv[0][0]
__________________________________________________________________________________________________
conv4_block3_1_relu (Activation (None, 14, 14, 256) 0 conv4_block3_1_bn[0][0]
__________________________________________________________________________________________________
conv4_block3_2_conv (Conv2D) (None, 14, 14, 256) 590080 conv4_block3_1_relu[0][0]
__________________________________________________________________________________________________
conv4_block3_2_bn (BatchNormali (None, 14, 14, 256) 1024 conv4_block3_2_conv[0][0]
__________________________________________________________________________________________________
conv4_block3_2_relu (Activation (None, 14, 14, 256) 0 conv4_block3_2_bn[0][0]
__________________________________________________________________________________________________
conv4_block3_3_conv (Conv2D) (None, 14, 14, 1024) 263168 conv4_block3_2_relu[0][0]
__________________________________________________________________________________________________
conv4_block3_3_bn (BatchNormali (None, 14, 14, 1024) 4096 conv4_block3_3_conv[0][0]
__________________________________________________________________________________________________
conv4_block3_add (Add) (None, 14, 14, 1024) 0 conv4_block2_out[0][0]
conv4_block3_3_bn[0][0]
__________________________________________________________________________________________________
conv4_block3_out (Activation) (None, 14, 14, 1024) 0 conv4_block3_add[0][0]
__________________________________________________________________________________________________
conv4_block4_1_conv (Conv2D) (None, 14, 14, 256) 262400 conv4_block3_out[0][0]
__________________________________________________________________________________________________
conv4_block4_1_bn (BatchNormali (None, 14, 14, 256) 1024 conv4_block4_1_conv[0][0]
__________________________________________________________________________________________________
conv4_block4_1_relu (Activation (None, 14, 14, 256) 0 conv4_block4_1_bn[0][0]
__________________________________________________________________________________________________
conv4_block4_2_conv (Conv2D) (None, 14, 14, 256) 590080 conv4_block4_1_relu[0][0]
__________________________________________________________________________________________________
conv4_block4_2_bn (BatchNormali (None, 14, 14, 256) 1024 conv4_block4_2_conv[0][0]
__________________________________________________________________________________________________
conv4_block4_2_relu (Activation (None, 14, 14, 256) 0 conv4_block4_2_bn[0][0]
__________________________________________________________________________________________________
conv4_block4_3_conv (Conv2D) (None, 14, 14, 1024) 263168 conv4_block4_2_relu[0][0]
__________________________________________________________________________________________________
conv4_block4_3_bn (BatchNormali (None, 14, 14, 1024) 4096 conv4_block4_3_conv[0][0]
__________________________________________________________________________________________________
conv4_block4_add (Add) (None, 14, 14, 1024) 0 conv4_block3_out[0][0]
conv4_block4_3_bn[0][0]
__________________________________________________________________________________________________
conv4_block4_out (Activation) (None, 14, 14, 1024) 0 conv4_block4_add[0][0]
__________________________________________________________________________________________________
conv4_block5_1_conv (Conv2D) (None, 14, 14, 256) 262400 conv4_block4_out[0][0]
__________________________________________________________________________________________________
conv4_block5_1_bn (BatchNormali (None, 14, 14, 256) 1024 conv4_block5_1_conv[0][0]
__________________________________________________________________________________________________
conv4_block5_1_relu (Activation (None, 14, 14, 256) 0 conv4_block5_1_bn[0][0]
__________________________________________________________________________________________________
conv4_block5_2_conv (Conv2D) (None, 14, 14, 256) 590080 conv4_block5_1_relu[0][0]
__________________________________________________________________________________________________
conv4_block5_2_bn (BatchNormali (None, 14, 14, 256) 1024 conv4_block5_2_conv[0][0]
__________________________________________________________________________________________________
conv4_block5_2_relu (Activation (None, 14, 14, 256) 0 conv4_block5_2_bn[0][0]
__________________________________________________________________________________________________
conv4_block5_3_conv (Conv2D) (None, 14, 14, 1024) 263168 conv4_block5_2_relu[0][0]
__________________________________________________________________________________________________
conv4_block5_3_bn (BatchNormali (None, 14, 14, 1024) 4096 conv4_block5_3_conv[0][0]
__________________________________________________________________________________________________
conv4_block5_add (Add) (None, 14, 14, 1024) 0 conv4_block4_out[0][0]
conv4_block5_3_bn[0][0]
__________________________________________________________________________________________________
conv4_block5_out (Activation) (None, 14, 14, 1024) 0 conv4_block5_add[0][0]
__________________________________________________________________________________________________
conv4_block6_1_conv (Conv2D) (None, 14, 14, 256) 262400 conv4_block5_out[0][0]
__________________________________________________________________________________________________
conv4_block6_1_bn (BatchNormali (None, 14, 14, 256) 1024 conv4_block6_1_conv[0][0]
__________________________________________________________________________________________________
conv4_block6_1_relu (Activation (None, 14, 14, 256) 0 conv4_block6_1_bn[0][0]
__________________________________________________________________________________________________
conv4_block6_2_conv (Conv2D) (None, 14, 14, 256) 590080 conv4_block6_1_relu[0][0]
__________________________________________________________________________________________________
conv4_block6_2_bn (BatchNormali (None, 14, 14, 256) 1024 conv4_block6_2_conv[0][0]
__________________________________________________________________________________________________
conv4_block6_2_relu (Activation (None, 14, 14, 256) 0 conv4_block6_2_bn[0][0]
__________________________________________________________________________________________________
conv4_block6_3_conv (Conv2D) (None, 14, 14, 1024) 263168 conv4_block6_2_relu[0][0]
__________________________________________________________________________________________________
conv4_block6_3_bn (BatchNormali (None, 14, 14, 1024) 4096 conv4_block6_3_conv[0][0]
__________________________________________________________________________________________________
conv4_block6_add (Add) (None, 14, 14, 1024) 0 conv4_block5_out[0][0]
conv4_block6_3_bn[0][0]
__________________________________________________________________________________________________
conv4_block6_out (Activation) (None, 14, 14, 1024) 0 conv4_block6_add[0][0]
__________________________________________________________________________________________________
conv5_block1_1_conv (Conv2D) (None, 7, 7, 512) 524800 conv4_block6_out[0][0]
__________________________________________________________________________________________________
conv5_block1_1_bn (BatchNormali (None, 7, 7, 512) 2048 conv5_block1_1_conv[0][0]
__________________________________________________________________________________________________
conv5_block1_1_relu (Activation (None, 7, 7, 512) 0 conv5_block1_1_bn[0][0]
__________________________________________________________________________________________________
conv5_block1_2_conv (Conv2D) (None, 7, 7, 512) 2359808 conv5_block1_1_relu[0][0]
__________________________________________________________________________________________________
conv5_block1_2_bn (BatchNormali (None, 7, 7, 512) 2048 conv5_block1_2_conv[0][0]
__________________________________________________________________________________________________
conv5_block1_2_relu (Activation (None, 7, 7, 512) 0 conv5_block1_2_bn[0][0]
__________________________________________________________________________________________________
conv5_block1_0_conv (Conv2D) (None, 7, 7, 2048) 2099200 conv4_block6_out[0][0]
__________________________________________________________________________________________________
conv5_block1_3_conv (Conv2D) (None, 7, 7, 2048) 1050624 conv5_block1_2_relu[0][0]
__________________________________________________________________________________________________
conv5_block1_0_bn (BatchNormali (None, 7, 7, 2048) 8192 conv5_block1_0_conv[0][0]
__________________________________________________________________________________________________
conv5_block1_3_bn (BatchNormali (None, 7, 7, 2048) 8192 conv5_block1_3_conv[0][0]
__________________________________________________________________________________________________
conv5_block1_add (Add) (None, 7, 7, 2048) 0 conv5_block1_0_bn[0][0]
conv5_block1_3_bn[0][0]
__________________________________________________________________________________________________
conv5_block1_out (Activation) (None, 7, 7, 2048) 0 conv5_block1_add[0][0]
__________________________________________________________________________________________________
conv5_block2_1_conv (Conv2D) (None, 7, 7, 512) 1049088 conv5_block1_out[0][0]
__________________________________________________________________________________________________
conv5_block2_1_bn (BatchNormali (None, 7, 7, 512) 2048 conv5_block2_1_conv[0][0]
__________________________________________________________________________________________________
conv5_block2_1_relu (Activation (None, 7, 7, 512) 0 conv5_block2_1_bn[0][0]
__________________________________________________________________________________________________
conv5_block2_2_conv (Conv2D) (None, 7, 7, 512) 2359808 conv5_block2_1_relu[0][0]
__________________________________________________________________________________________________
conv5_block2_2_bn (BatchNormali (None, 7, 7, 512) 2048 conv5_block2_2_conv[0][0]
__________________________________________________________________________________________________
conv5_block2_2_relu (Activation (None, 7, 7, 512) 0 conv5_block2_2_bn[0][0]
__________________________________________________________________________________________________
conv5_block2_3_conv (Conv2D) (None, 7, 7, 2048) 1050624 conv5_block2_2_relu[0][0]
__________________________________________________________________________________________________
conv5_block2_3_bn (BatchNormali (None, 7, 7, 2048) 8192 conv5_block2_3_conv[0][0]
__________________________________________________________________________________________________
conv5_block2_add (Add) (None, 7, 7, 2048) 0 conv5_block1_out[0][0]
conv5_block2_3_bn[0][0]
__________________________________________________________________________________________________
conv5_block2_out (Activation) (None, 7, 7, 2048) 0 conv5_block2_add[0][0]
__________________________________________________________________________________________________
conv5_block3_1_conv (Conv2D) (None, 7, 7, 512) 1049088 conv5_block2_out[0][0]
__________________________________________________________________________________________________
conv5_block3_1_bn (BatchNormali (None, 7, 7, 512) 2048 conv5_block3_1_conv[0][0]
__________________________________________________________________________________________________
conv5_block3_1_relu (Activation (None, 7, 7, 512) 0 conv5_block3_1_bn[0][0]
__________________________________________________________________________________________________
conv5_block3_2_conv (Conv2D) (None, 7, 7, 512) 2359808 conv5_block3_1_relu[0][0]
__________________________________________________________________________________________________
conv5_block3_2_bn (BatchNormali (None, 7, 7, 512) 2048 conv5_block3_2_conv[0][0]
__________________________________________________________________________________________________
conv5_block3_2_relu (Activation (None, 7, 7, 512) 0 conv5_block3_2_bn[0][0]
__________________________________________________________________________________________________
conv5_block3_3_conv (Conv2D) (None, 7, 7, 2048) 1050624 conv5_block3_2_relu[0][0]
__________________________________________________________________________________________________
conv5_block3_3_bn (BatchNormali (None, 7, 7, 2048) 8192 conv5_block3_3_conv[0][0]
__________________________________________________________________________________________________
conv5_block3_add (Add) (None, 7, 7, 2048) 0 conv5_block2_out[0][0]
conv5_block3_3_bn[0][0]
__________________________________________________________________________________________________
conv5_block3_out (Activation) (None, 7, 7, 2048) 0 conv5_block3_add[0][0]
__________________________________________________________________________________________________
avg_pool (GlobalAveragePooling2 (None, 2048) 0 conv5_block3_out[0][0]
__________________________________________________________________________________________________
dense_1 (Dense) (None, 1) 2049 avg_pool[0][0]
==================================================================================================
Total params: 23,589,761
Trainable params: 23,536,641
Non-trainable params: 53,120
__________________________________________________________________________________________________