MIMIC-III and Clinical Outcomes
Mortality is the most commonly used outcome in prediction studies in critical care, and in fact potentially across all of medicine. Since it is a strong surrogate for realness, machine learning practitioners use this signal to infer the relationship between clinical data and patient outcome. Mortality is a deceptively simple outcome to define. In fact, the nature of retrospective data often complicates matters. For example, in MIMIC database, a number of patients have two consecutive hospitalizations in which they die as organ donors. These consecutive hospitalizations are treated as distinct hospital admissions.
However, the underlying reason is the same. Mortality is susceptible to selection bias based on the source of the information. For example, many studies report hospital mortality as it is feasible to collect this information. However, the number of deaths recorded in the hospital database might not include the number of people that have died after they have been discharged and went to the home care. This adds noise to the outcome.
Other important factors in defining mortality as an outcome are controllable by the researcher. For example, defining mortality as death within 30 days of admission will provide a stronger signal for immediate physiological abnormality, which is likely related to the patient’s admission. On the other hand, one-year mortality will emphasize chronic illness in parallel conditions.
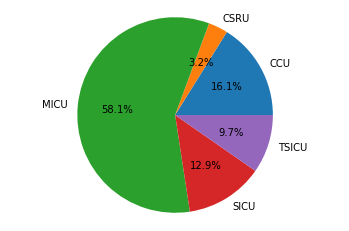
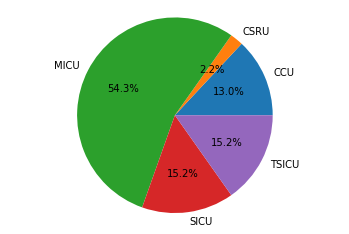
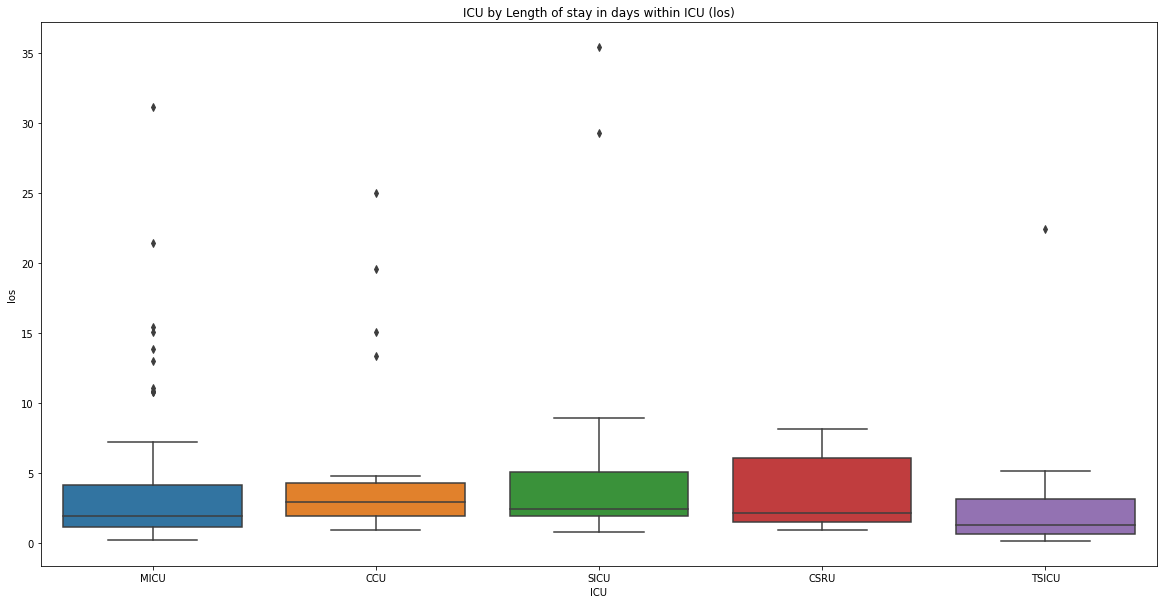
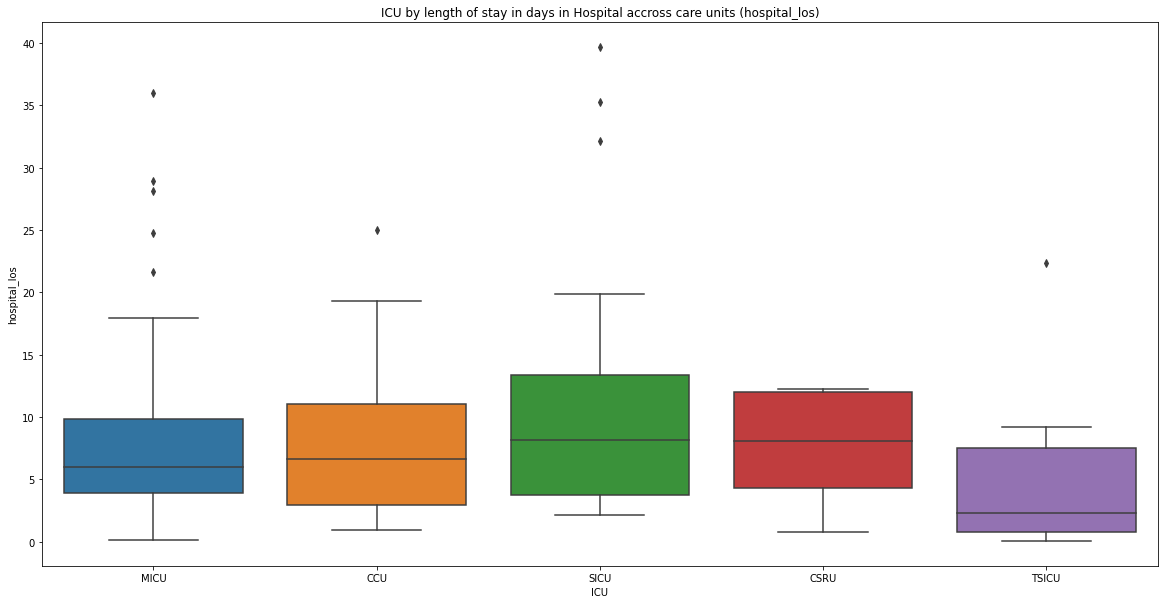
in this article we will see how we can extract the mortality numbers of adult patients who were admitted to the ICU and the distribution of this mortality numbers across the different ICUs in MIMIC dataset. If a patient’s death was registered while the patient was in ICU, or six hours before being admitted to, or six hours after leaving the ICU, we will assume that the patient has died in the intensive care unit.
Subscribe